By Aniket Gambhire, Associate Data Scientist at AlgoAnalytics
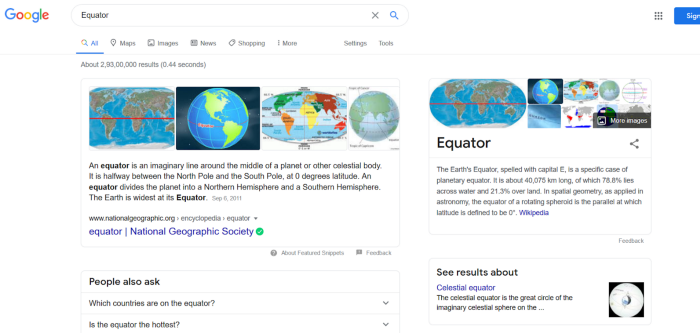
Have you ever wondered how when you search for something or someone in the Google search engine you see relevant stories, Wikipedia, and images, etc. For example, look at the search results for “Equator” in Figure 1.
How is all this information identified and displayed correctly? That’s the “Knowledge Graph” at work. Google stores all the information related to the Equator in the Knowledge Graph database and when you do a query it will pop up everything.
What exactly is a Knowledge Graph?
A Knowledge Graph acquires and integrates information into an ontology and applies a reasoner to derive new knowledge. Or in simple terms, Knowledge Graphs are large networks of entities, their semantic types, properties, and relationships between entities. [1]
To understand this better through an example, let’s see the Knowledge Graph shown in Figure 2.
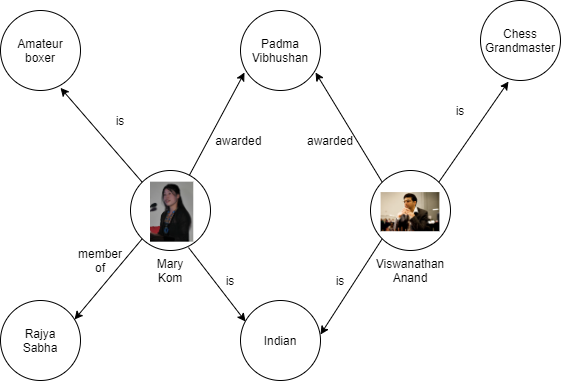
Figure 2: Example of a Knowledge Graph
Here, Figure 2 shows all the interconnected information with Mary Kom, that she is an Indian amateur boxer, a member of Rajya Sabha, and was awarded the Padma Vibhushan.
Knowledge Graph uses the triples to describe facts present in the real world. A triple is a tuple of subject, predicate, and object where subject and object are entities and predicate is the relationship between entities. (Mary Kom, is, Amateur Boxer) is a triple present in Figure 2.
With the help of a Knowledge Graph, we can transform unstructured data into structured ways and extract knowledge from it to solve some important problems.
As a case study, let’s look at our work at Algoanalytics, where we built a knowledge graph from medical discharge summaries to find all medical information related to patients. We named it the “Patient Knowledge Graph”.
The high-level design of Patient Knowledge Graph is illustrated in Figure 3.
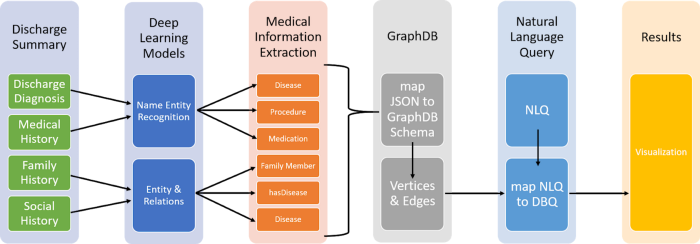
Figure 3: High-Level Design
We built the Knowledge Graph by using the text of discharge summaries from the hospital database of MIMIC Physionet Org. And followed below steps-
1. Text Extraction
Before extracting text from discharge summaries let’s understand the structure of it. Typically, all discharge summaries are divided into four sections which are family history, social history, medical history, and discharge diagnosis, and each section shows unique facts related to the patient. Each section has a fixed pattern in the discharge summary. Therefore, we extracted text from them using regular expressions.
2. Entities and Relation Extraction
After extracting the four sections, let’s extract entities and relations from each section to create triples.
Family History — Contains disease information of the patient’s family members from the maternal and paternal side (if they had any disease) and whether family members are alive or dead. The entities present in the family history are individual family members, associated diseases, and death related terms such as dead, expired, departed, death, deaths, dies, dying, passed. And we defined two relations as “hasDisease” for all diseases and “is” for all death-related terms. Now we have to prepare triples to build a knowledge graph where the subject is family member, the object is disease/death term and the predicate is the relation we defined earlier. We created a list of all family relationships to cover all possible triples. To extract disease and death term entities we used scispacy which is a python package containing spaCy models for processing biomedical, scientific or clinical text.
By iterating over all discharge summaries we created a list of diseases and death related terms using scispacy. Further, using a list of family relationships and diseases we built a training file of a triple with “hasDisease” and “is” relation which is used to build knowledge embedding using TransR model of OpenKE. Using this knowledge embedding we trained the ERNIE relation classification model to predict the relation between family relationships and disease entities.
Social History —Provides information about a patient’s social life, such as if he/she is married or single, his/her profession, his/her habits such as smoking or drinking etc. As the social history section contains the general information of the patient, we have used Stanford Open Information Extraction 5.1 to extract triple from the text. Here, for segmentation, we have used spaCy an NLP library.
Medical History —Provides information of the disease the patient had in the past and sometimes the reason for his/her illness.
Discharge Diagnosis —Contains information on the diagnoses or procedures done on the patient during hospitalization.
The medical terms of our interest in both Medical History and Discharge Diagnosis sections are the name of the diseases, procedures. Hence, here one entity is disease name or procedure name, where the disease is extracted using scispacy and the procedure is extracted using BioBERT. And the other entity is the patient themself. The relation is “hasDisease” and “hasProcedure” which are defined manually.
3. Build Knowledge Graph
After extracting the entities and relations from text, all of this information is stored in the JSON file of their respective sections. In our project, we have used the OrientDB graph database to copy information from all these JSON files by using database schemas. We have built a knowledge graph of vertices and edges as shown in Figure 4.
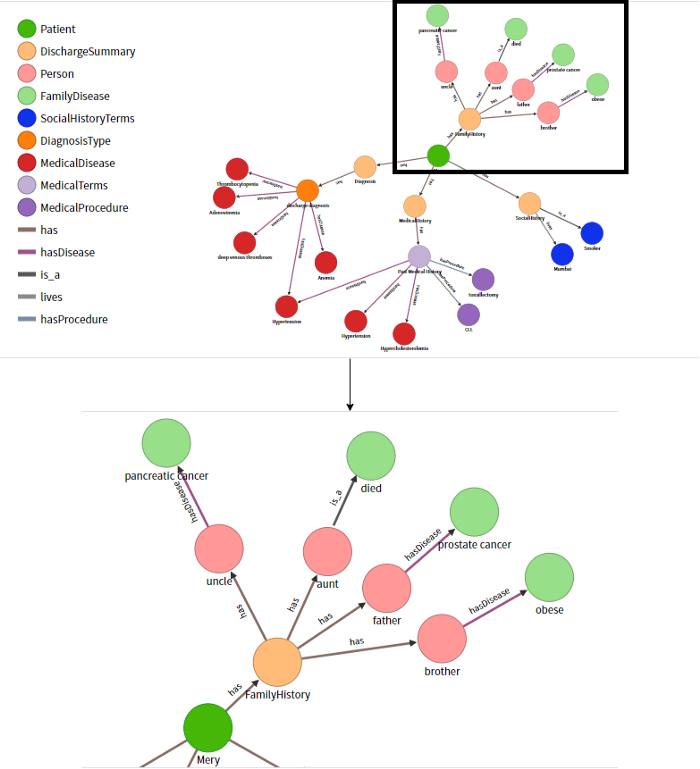
Figure 4: Knowledge Graph for Patient Mery
4. Information Retrieval and Visualization
The Knowledge Graph is created and stored in the OrientDB graph database. We can visualize it in the browser with the help of the OrientDB Studio as shown in Figure 4.
Using OrientDB query language, we can retrieve the large scale knowledge from the graph. Some Natural Language queries used to retrieve knowledge related to patients are listed below-
- What are the diseases associated with patients who smoke?
- How many patients with disease <disease name> AND the family history of disease <disease name>?
- What are the diseases for patients with a family history of disease <disease name>?
The Patient Knowledge Graph is helpful in the medical domain for doctors to see the patient details pictorially instead of going through the whole discharge summary line by line. The doctors can use it as a question answering system to extract information with one click. The medical domain knowledge graph can also be used in electronic health records, health risk prediction, and for clinical decision support.
Not just in the medical domain (demonstrated in the above case study), but the Knowledge Graph has innumerable applications in a variety of domains and industries. For example, in the finance industry, Knowledge Graph is used for knowledge management, automated fraud detection, and data governance. It is also used in community-driven knowledge sharing applications such as Wikidata.
Demo and Contact Info:
For demo visit our link https://algoanalytics.com/demoapp
For further information, please contact: info@algoanalytics.com
References:
[1] Towards a Definition of Knowledge Graphs
URL: http://ceur-ws.org/Vol-1695/paper4.pdf
[2] A Survey on Knowledge Graphs: Representation, Acquisition and Applications
URL: https://arxiv.org/pdf/2002.00388.pdf
[3] A Review of Relation Machine Learning for Knowledge Graphs
URL: https://arxiv.org/pdf/1503.00759.pdf
[4] Open Information Extraction 5.1 System
URL: https://github.com/dair-iitd/OpenIE-standalone
[5] ScispaCy github
URL: https://allenai.github.io/scispacy/
[6] Open Source Framework for Knowledge Embedding
[7] ERNIE: Enhanced Language Representation with Informative Entities

